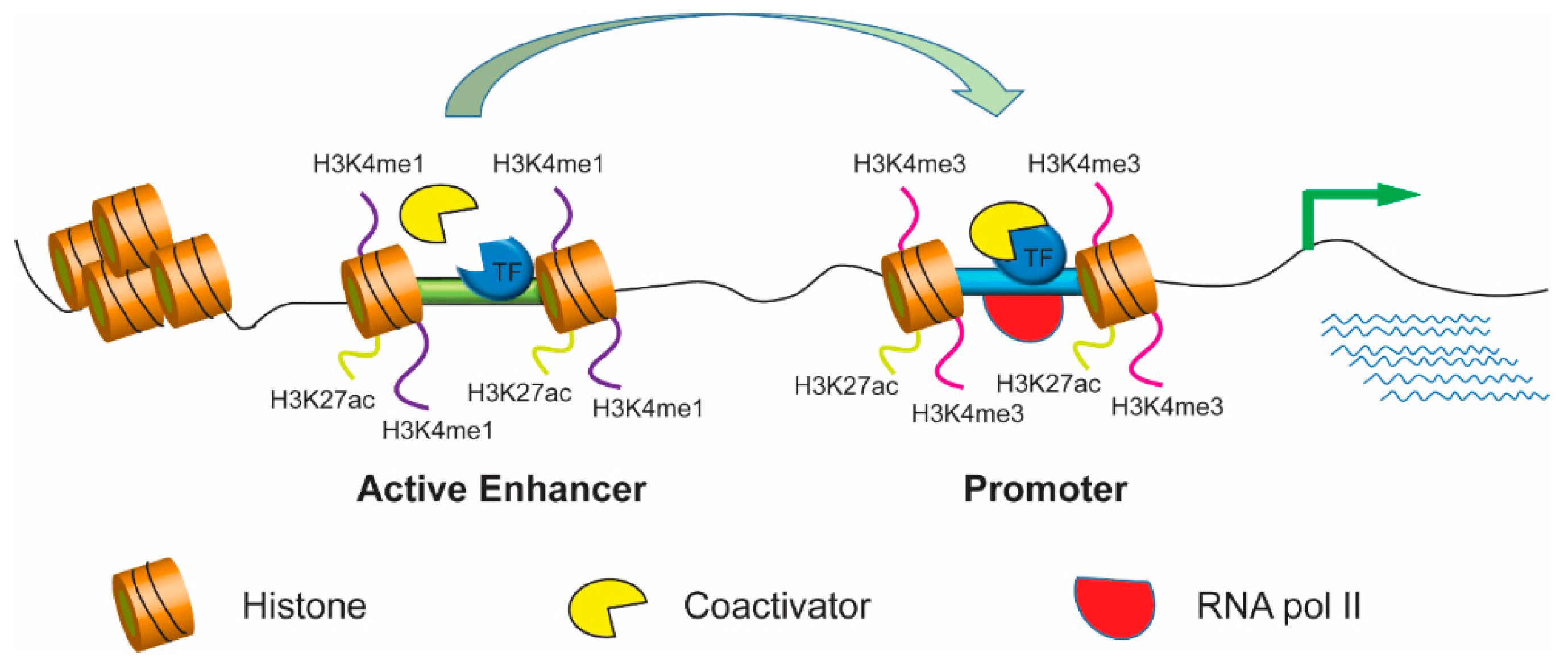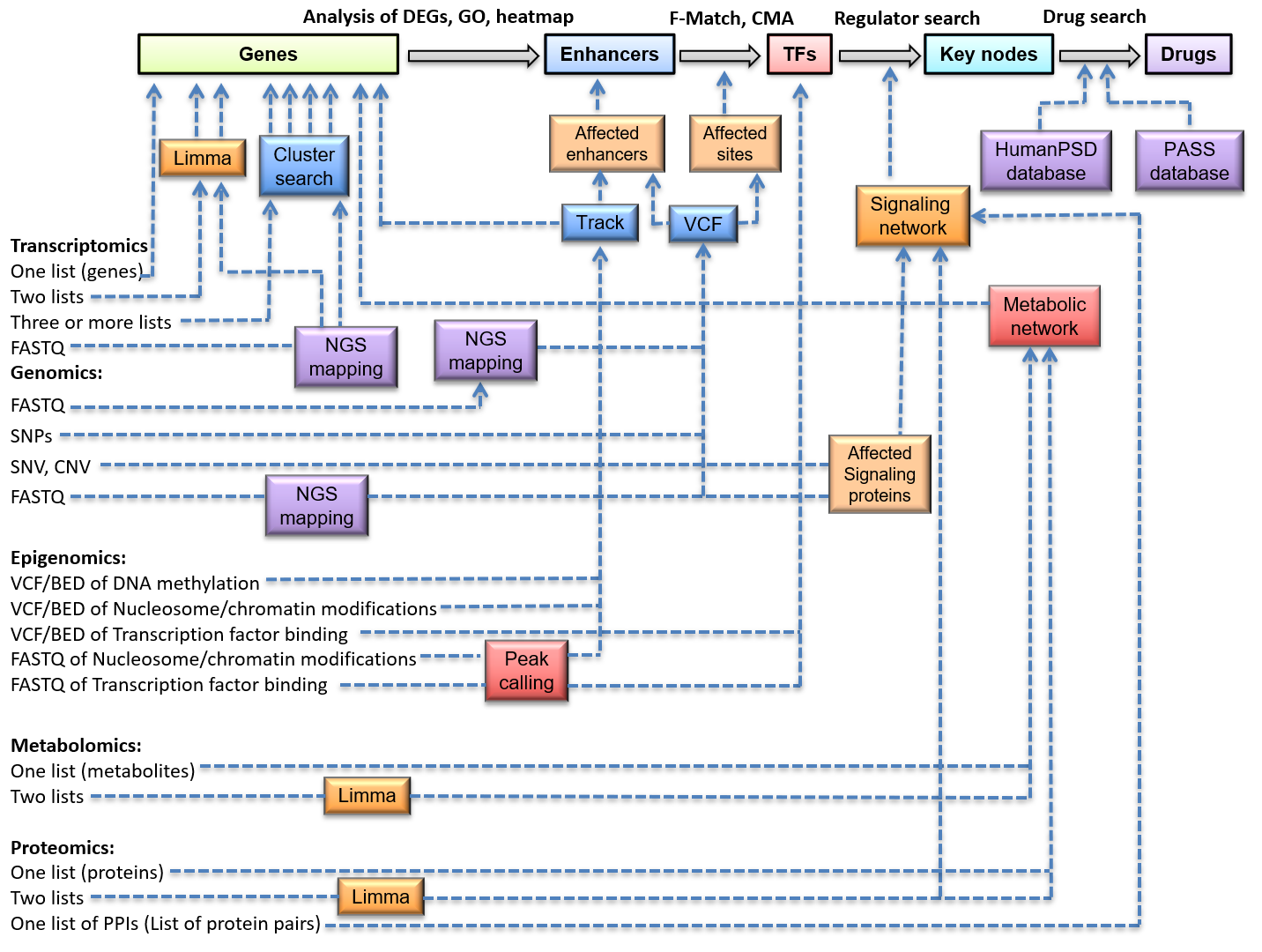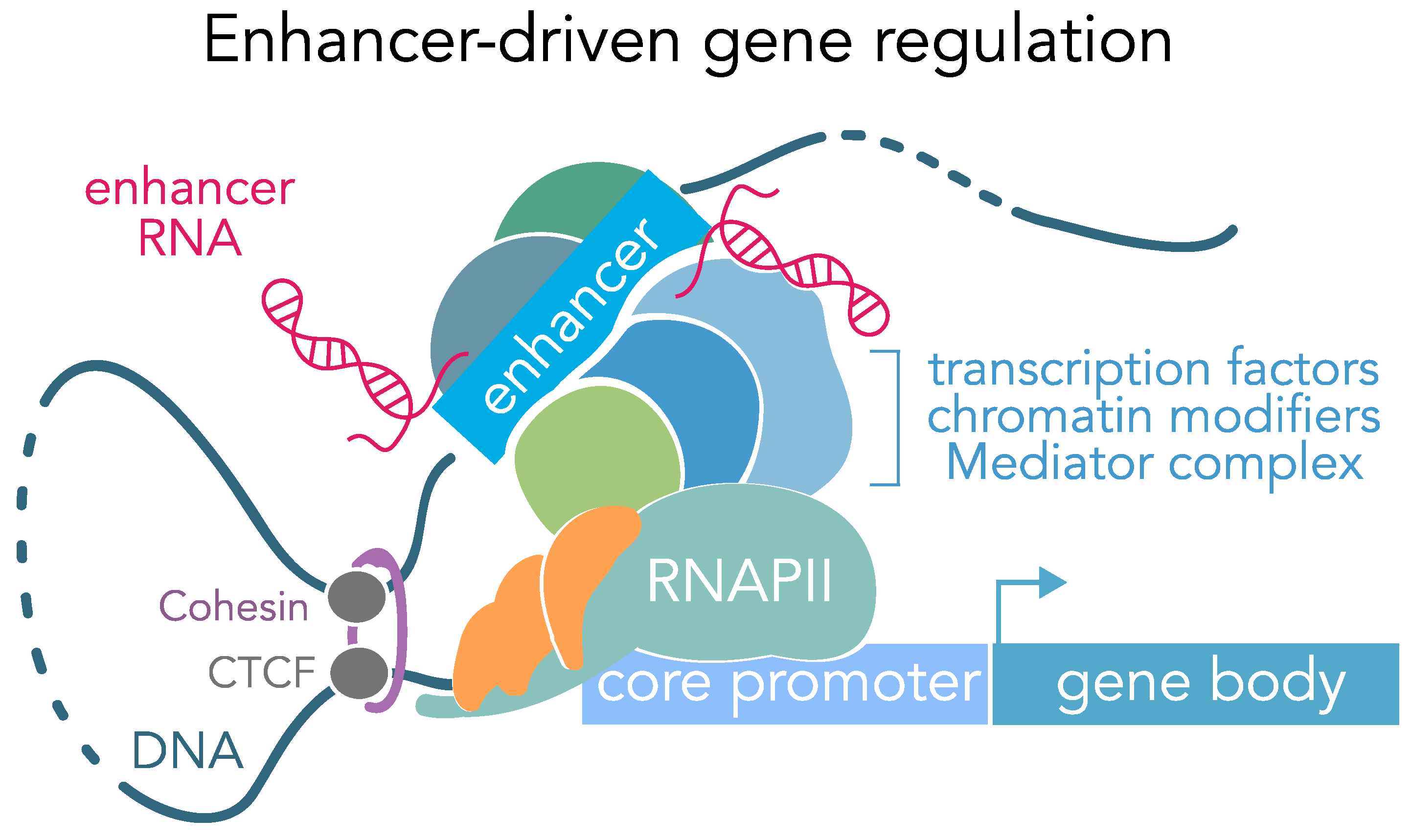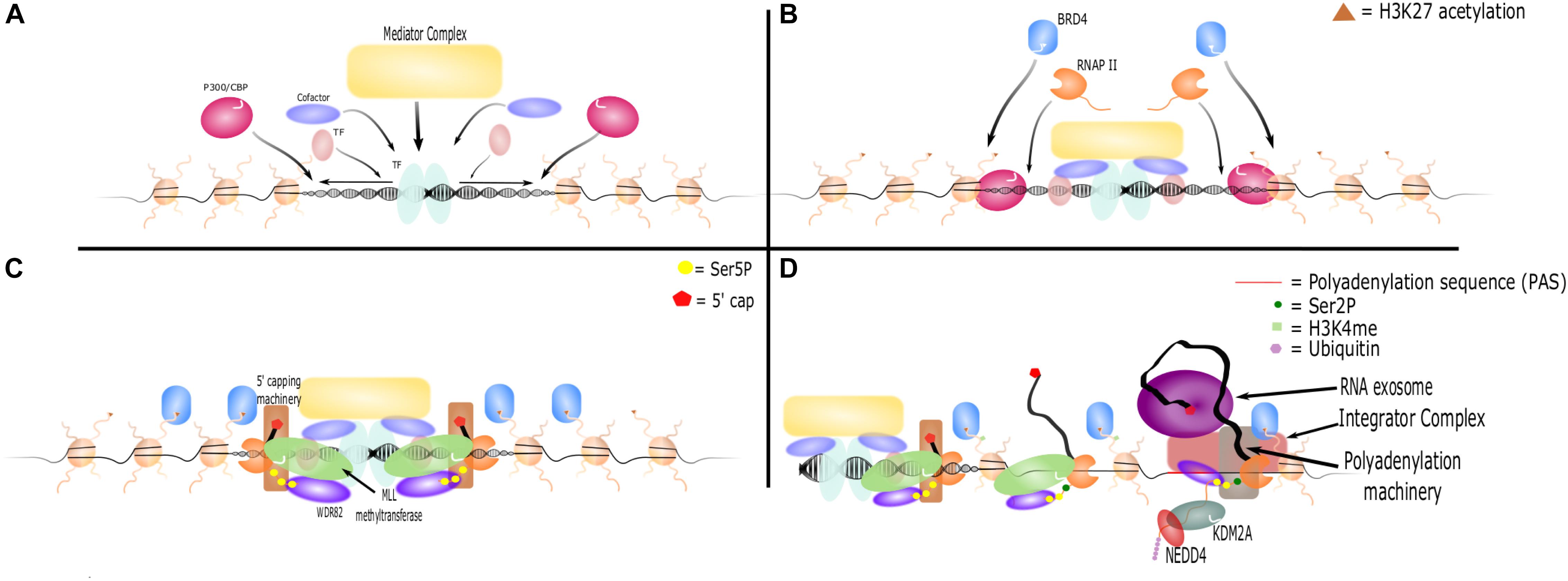
Frontiers | Diversity and Emerging Roles of Enhancer RNA in Regulation of Gene Expression and Cell Fate

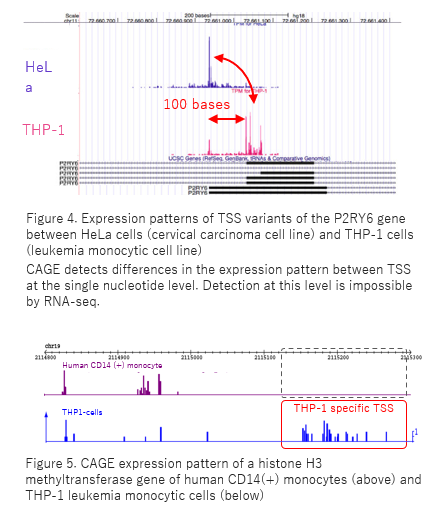
NET-CAGE characterizes the dynamics and topology of human transcribed cis-regulatory elements | Nature Genetics

De novo identification of transcribed enhancers and super-enhancers in... | Download Scientific Diagram

A High-Resolution Map of Human Enhancer RNA Loci Characterizes Super- enhancer Activities in Cancer - ScienceDirect

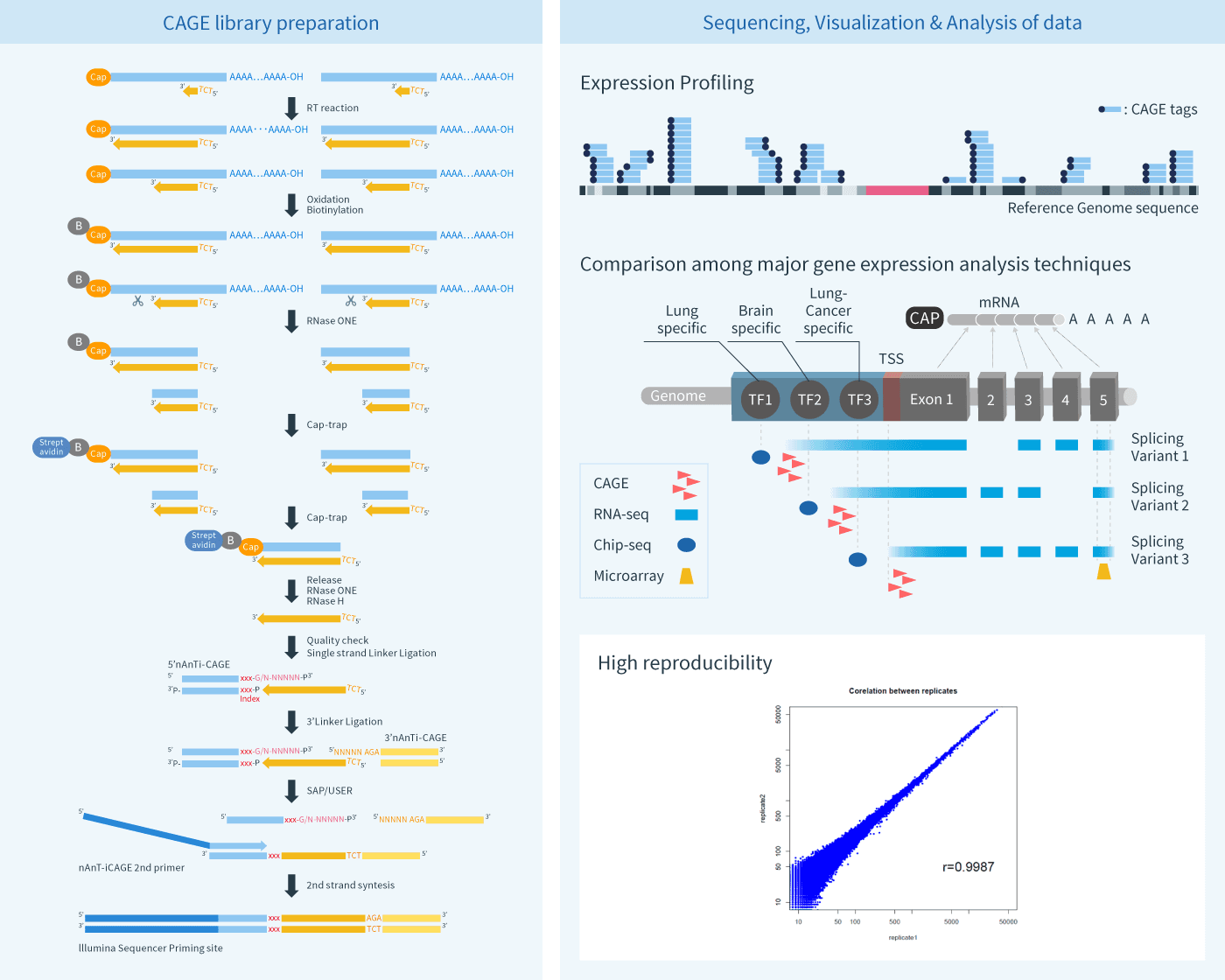
C1 CAGE detects transcription start sites and enhancer activity at single-cell resolution | Nature Communications

Enhanced Identification of Transcriptional Enhancers Provides Mechanistic Insights into Diseases: Trends in Genetics

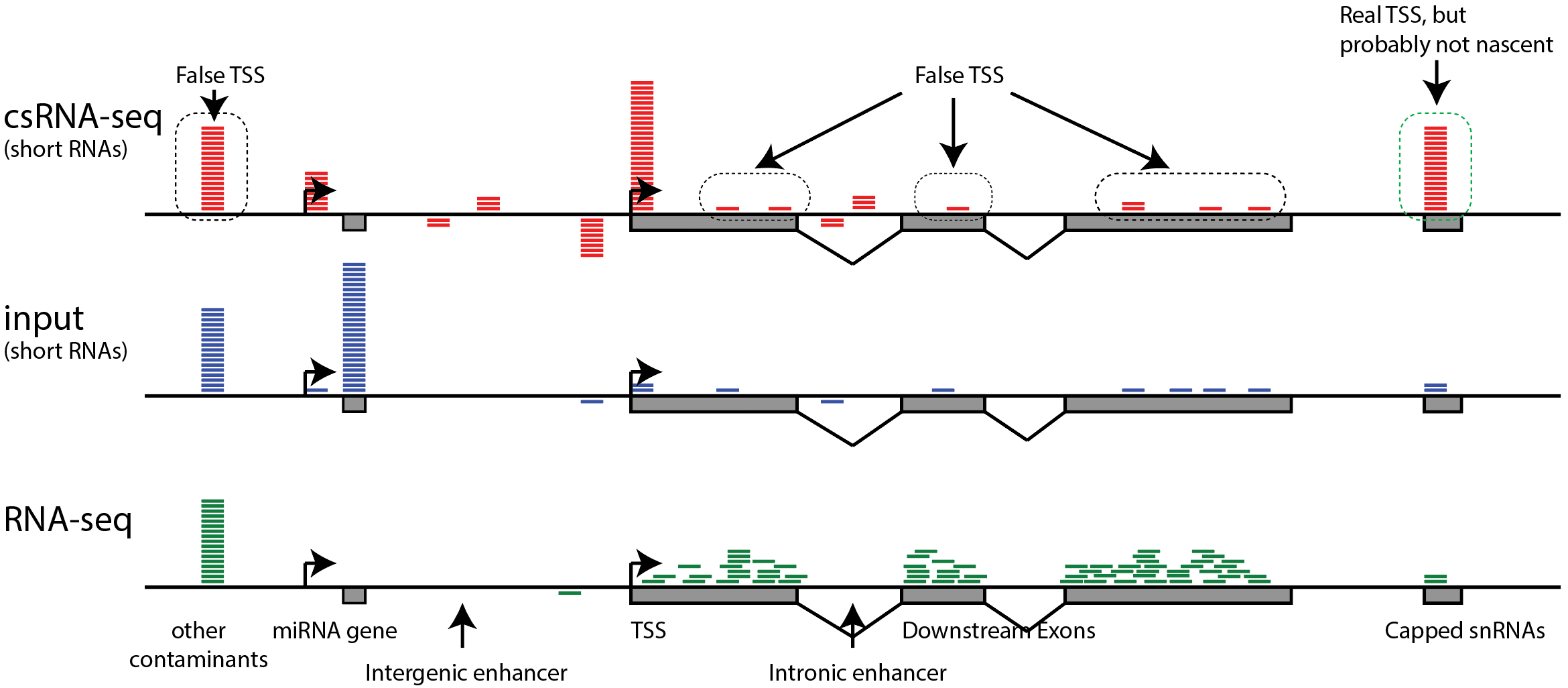
Nascent RNA sequencing analysis provides insights into enhancer-mediated gene regulation | BMC Genomics | Full Text

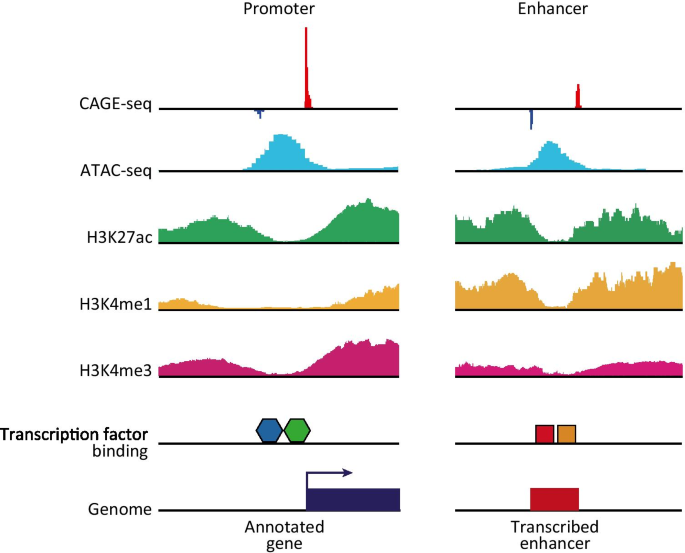
Altered Enhancer and Promoter Usage Leads to Differential Gene Expression in the Normal and Failed Human Heart | Circulation: Heart Failure

A neural network based model effectively predicts enhancers from clinical ATAC-seq samples | Scientific Reports
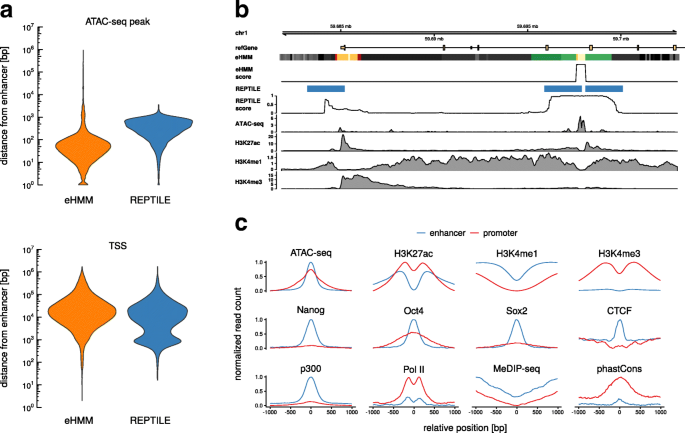
Predicting enhancers in mammalian genomes using supervised hidden Markov models | BMC Bioinformatics | Full Text
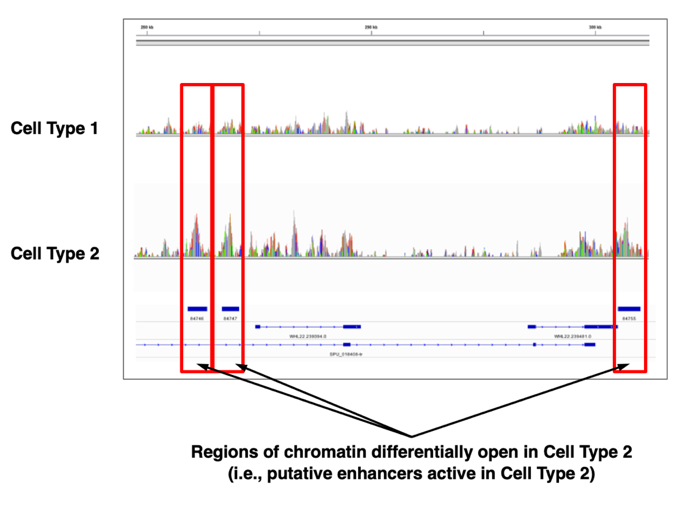
Genomic Feature Datasets for Analysis of Regulatory DNA Elements – Resource for Developmental Regulatory Genomics